
I grew up in a bushy outer suburb of Melbourne and spent most of my spare time looking for animals in my backyard and down at the local creek. As my travels grew wider, I began to find the geographic distributions of species fascinating. What characteristics of a species allow it to live in the harshest of the Australian deserts? What is it about a montane specialist that prevents it from living at low elevation?
These questions—and a general curiosity about species, their traits and the environments they live in—motivate my research and have steered me towards working with the Atlas of Living Australia.
The limits of species distribution mapping
The ALA provides detailed information on Australian species’ distributions. But we actually have very limited understanding of why the distributions appear as they do. This limits our ability to anticipate the geographic spread or decline of species as the Australian environment changes.
Understanding the causes of species distribution limits is not a trivial problem. Ecological systems are a complex web of interactions. The processes limiting species distribution and abundance may occur over vast spatial and temporal scales. Today, we have detailed quantitative information on climate and terrain. We can use this data to statistically model species occurrences as a function of environmental factors. This is often referred to as ‘ecological niche modelling’, and there are tools to do this in the ALA. However, the current models are limited in what they can say about the underlying processes, and about what might happen in new scenarios.
Introducing traits: connecting functional characteristics of a species with its environment
Instead of starting with occurrence records, and statistically relating them to environmental data, another approach is to build models that explicitly represent the cause-and-effect links between the functional traits of organisms and their environments. This is the path I have been taking in my research, something I have called ‘mechanistic niche modelling’.
I knew that building these models on the basis of physical first-principles would give the greatest predictive power and generality across species and environments. Although I didn’t know a lot of physics when I started, I was able to follow the trail blazed by a number of intrepid ecologists (especially Prof. Warren Porter, Department of Integrative Biology, University of Wisconsin and Prof. Bas Kooijman, Department of Theoretical Biology, Vrije University, Amsterdam) who had applied the fundamental laws of thermodynamics to explore how organisms exchange energy and matter with their environments. This field is called ‘biophysical ecology’.
The equations of biophysical ecology allow us to ask: Will an organism be warm enough to forage? Will it overheat and die? How does the environment affect its energy requirements? How much water will it lose, and is that balanced by what it is getting from food? How long will it take to hatch, mature and reproduce? Will it die before getting a chance to reproduce?
Answers to these questions can be put together to understand what limits species distribution and abundance. This approach can also be used to plan or interpret field surveys, allowing us to determine what time of day or year we are most likely to find a given species in a particular environment.
Our results so far

To understand how climate limits the distribution, life histories and activity patterns of a broad range of species, I developed a suite of models for making biophysical calculations with an R package, NicheMapR.
I used these models in our recent study of the Sleepy Lizard Field tests of a general ectotherm niche model show how water can limit lizard activity and distribution. This study showed that Sleepy Lizard distribution is far more constrained by water than by temperature, and that it will become inactive to avoid water loss during dry spells.
We have also analysed the water requirements of the endangered Night Parrot (Pezoporus occidentalis) in the paper An estimate of the water budget for the endangered night parrot of Australia under recent and future climates. This revealed that, to survive, the night parrot must have access to standing water during the summer. This has important implications for managing the resources of this critically endangered species.
Looking forward – a world with more trait data
The tools now exist to answer fundamental questions about how species are limited by their environments, but what we need most is the trait data. Which traits we need is very clear – the equations of ‘biophysical models’ precisely define them.
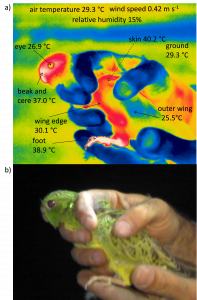
For the models of heat exchange, water balance, and potential activity time, we need traits like solar reflectance, resistance to water loss, surface area, insulation (e.g. fur depth), temperature and desiccation thresholds for activity and survival, and the water content of the body, food and faeces.
For the metabolic life cycle model, we need observations such as development time, reproduction rates, metabolic rate, lengths and weights at life cycle transitions (birth, maturation, ultimate size): from these observations a set of 12 parameters are estimated that are the same for every species.
Trait data and the ALA
My vision is to develop a database of these mechanistic niche modelling traits in the ALA.
The idea would be that modelling packages (like NicheMapR) can directly query the ALA for the relevant traits, in just the same way that correlative species distribution modelling packages query the ALA for occurrence records.
Although traits are not currently stored on the ALA, they are interested in incorporating trait data into their repository, and there are other research groups keen to see this happen. There are many different ways to use traits in biological analyses, and an almost infinite range of possibilities for what traits to include and how to specify them. The advantage of starting with the traits required for mechanistic niche modelling is that they are a clearly defined set, varying among taxa in a limited way (e.g. feather/fur depth is not required for lizards). And, although these traits are tightly defined for a specific purpose, their fundamental biological importance makes them useful for all sorts of other questions.
Building this trait database on the ALA would create a step change in our ability to compute the climate responses of living Australia, and thus in our ability to conserve and manage our biodiversity in the environments to come.
More information
- I made the suite of models for biophysical calculations with an R package, NicheMapR. Please contact me, if you would like further information.
- For more information on traits and the ALA, contact Hamish Holewa or Michael Hope on support@ala.org.au .