PhyloJive (Phylogeny Javascript Information Visualiser and Explorer) is a web based application that places biodiversity information aggregated from many sources onto compact phylogenetic trees. PhyloJive:
- is entirely client-side
- renders in the web browser on a HTML5 canvas
- requires no plugins
- is open source
- works in current versions of all major browsers
- is built on the InfoVis JavaScript library
The project is the brainchild of Garry Jolley-Rogers and Joe Miller and was developed by Temi Varghese and Garry Jolley-Rogers as part of the Taxonomy Research & Information Network (TRIN) – see the original project page, original code repository and ALA code repository. The ALA has contributed to the PhyloJive codebase to integrate a number of web services: occurrence data, maps and character data from Identify Life. This work has been undertaken with help and advice from Joe Miller.
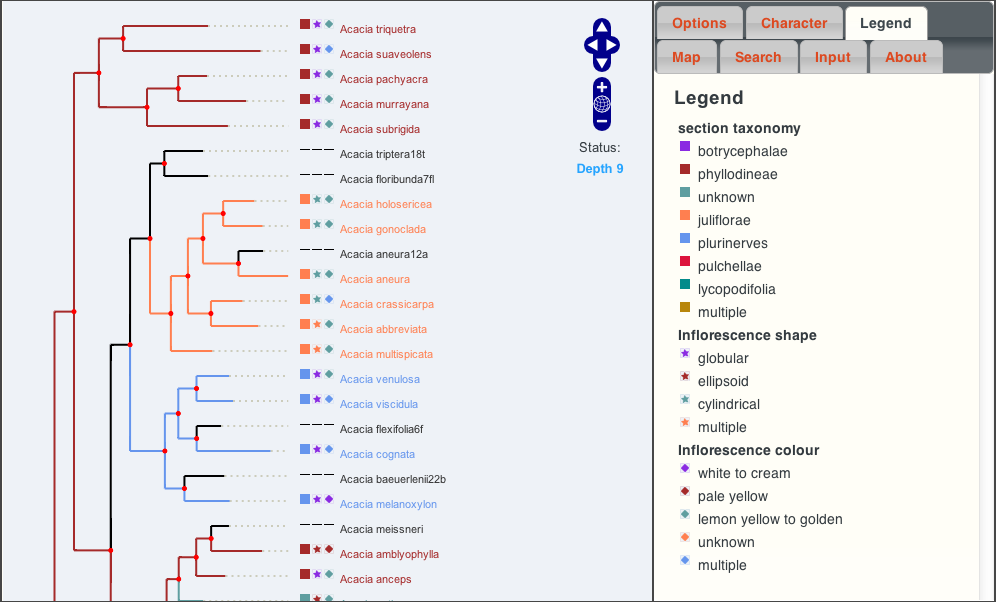
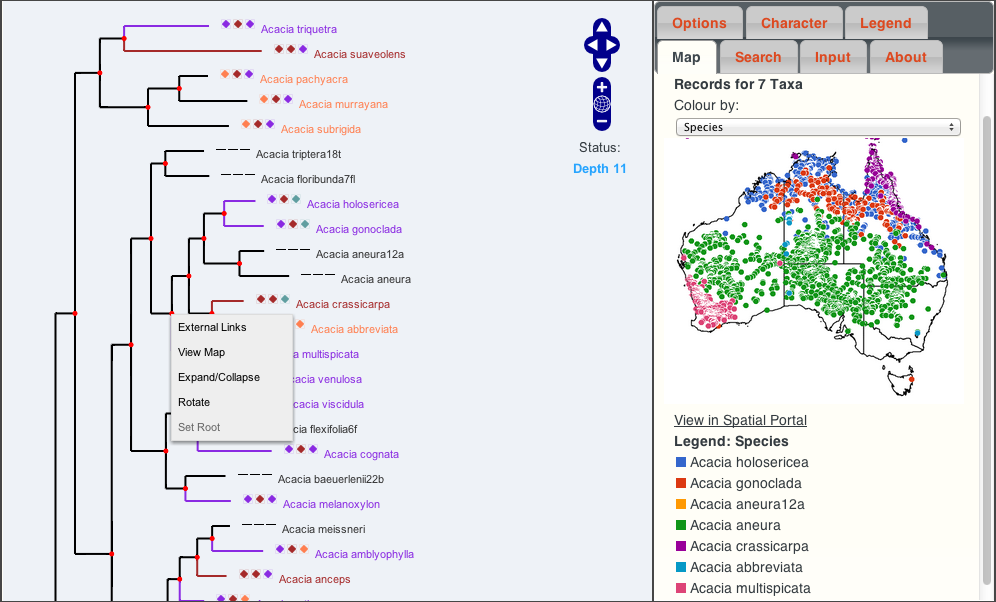
The following screen shot shows a phylogenetic tree for the Acacia genus.
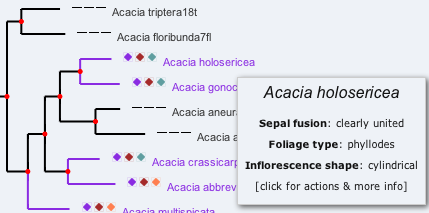
The coloured icons next to the scientific names represent different values for the three “characters” selected (these can be changed in the “character” tab), as shown in the legend panel on the right. Tree branches (and nodes for common ancestors) are coloured according to the predicted value for the first (chosen) character, as calculated using reverse parsimony.
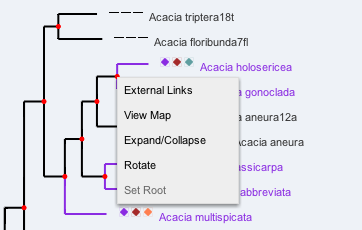
Mousing-over a node or name shows the names of the selected characters and their values. Clicking on a node or name brings up a menu to allow the user to perform a number of tasks.
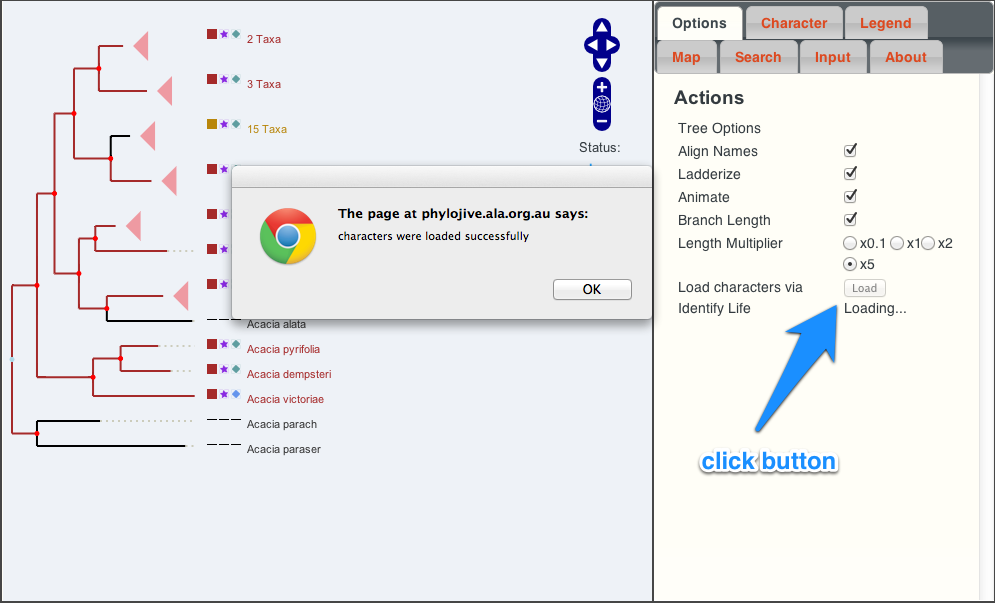
Importing characters from Identify Life
The ability to import characters from Identify Life is illustrated below. In the “options” tab, click the button labelled “Load” (currently only available for a small number of taxa groups). Once completed, the new characters can seen in the “Character” tab via the drop-down menu.
Mapping
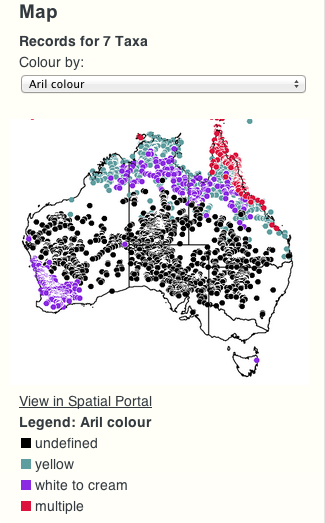
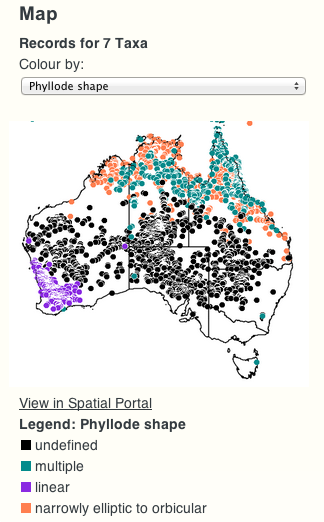
Another new feature is the ability to view a map of occurrence records for a given taxon or ancestral node (map will display all the “leaf” taxa for that node).
Furthermore, the colouring of the dots on the map can be changed from the (default) “by species” to any character available on the tree. In this way it is possible to get a geographic representation of the different possible character states (values).
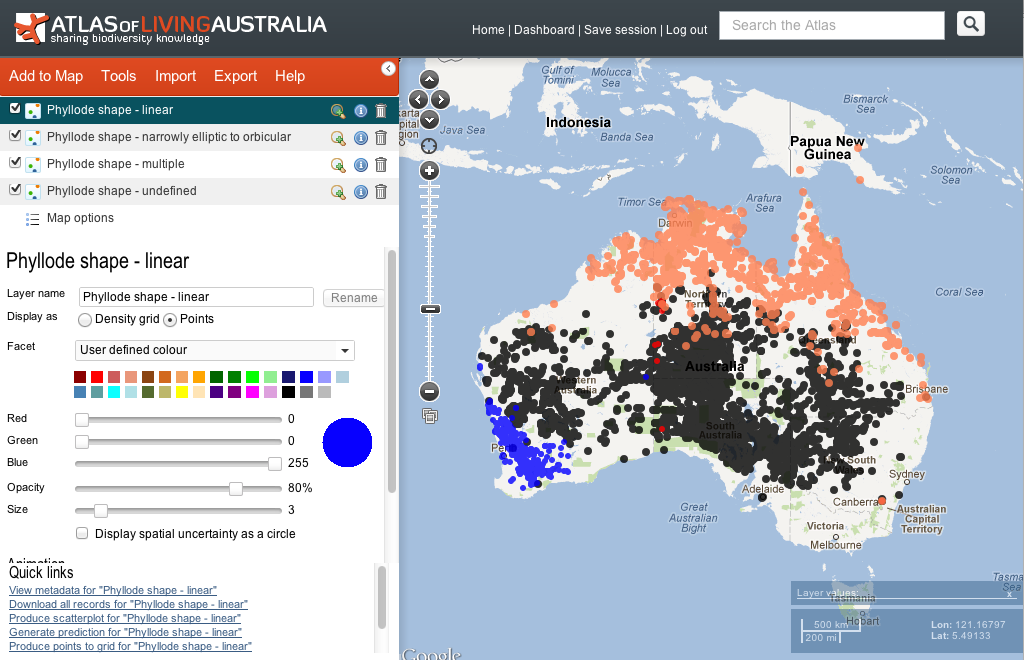
Lastly, these maps can be the basis of a spatial analysis by automatically importing the different colour schemes as separate spatial layers in the ALA spatial portal. Clicking the link “View in Spatial Portal” opens the ALA Spatial Portal in a new window (shown below). Layers can be hidden, combined, filtered by other criteria (e.g. collector, institution, etc) and various environmental layers can be overlaid or used as the basis of modelling, etc.
These are just a few of the features in PhyloJive. A beta version is available at http://phylojive.ala.org.au/.